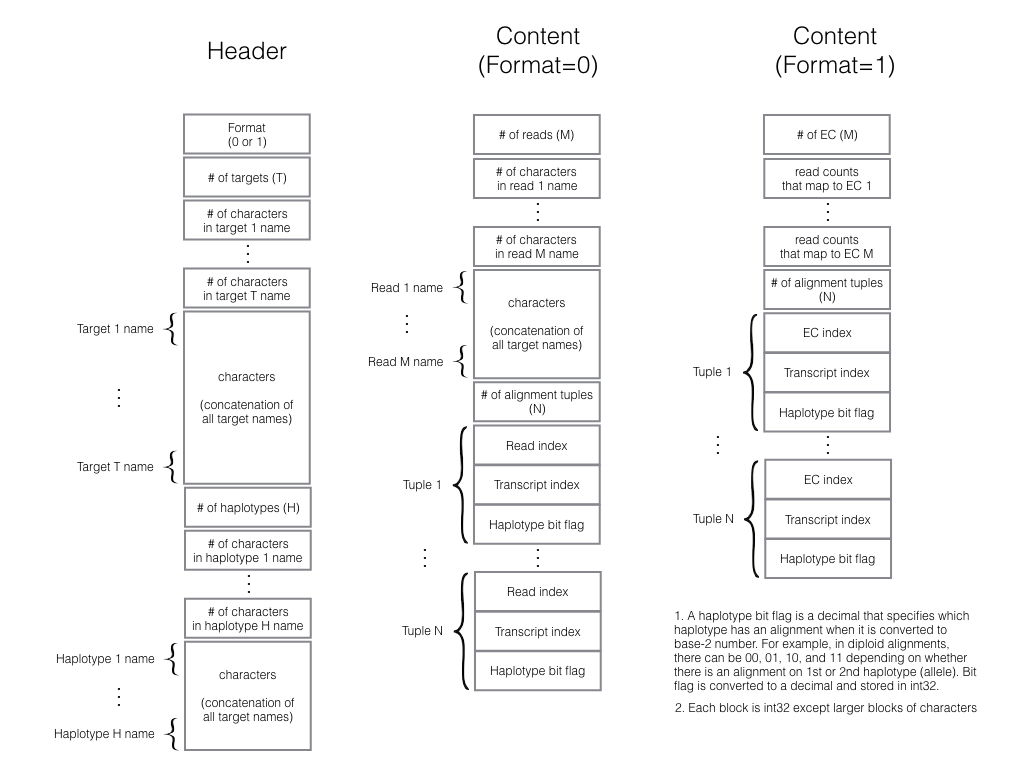
kallisto-align runs kallisto, processes its internal data object into an alignment incidence matrix, and exports the matrix in a pre-defined binary format for use with other tools such as emase or emase-zero. It is a companion utility for our bigger project that aims to provide a suite of tools to compare, contrast, or combine the results of different alignment strategies as envisioned in our alntools.
- Free software: GPLv3 license
Requirements
CMake version >= 2.8.12
HDF5 C library
zlib
Installation
To build kallisto-align you simply clone and compile.
$ git clone https://github.com/churchill-lab/kallisto-align.git
$ cd kallisto-align
$ cmake .
$ make
Note: kallisto will be downloaded and compiled for you and be located at:
external/src/kallisto-build/src/kallisto.
You can now either keep the kallisto and kallisto-align binaries where they are or move them to a more suitable location, e.g. /usr/local/bin.
Usage
We first need to build a kallisto index file. For example, given our target sequences are in a fasta file transcriptome.fa:
$ kallisto index -i transcriptome.idx transcriptome.fa
Please refer to the kallisto manual for more options.
Once the index file is created, you can run kallisto-align:
kallisto-align [OPTIONS...]
Options:
-f, --file READ FILE Input read file name
-i, --index INDEX FILE Input index file name
-b, --bin OUTPUT FILE Output file name
--reads Toggle read-level binary format (Format=0), else Equivalence Classes
-l, --load View the binary file
--help Print this message and exit
For example,
$ kallisto-align -f my_sample.fastq -i transcriptome.idx -b my_sample.bin
The binary file can be converted into emase format with alntools. It can also be used directly with emase-zero.
Output